Accumulation of body fat engenders increased risk and predisposition for adverse treatment outcome for many disease conditions including heart disease, diabetes mellitus, metabolic disorders, obstructive sleep apnea, certain lung disorders, and certain cancers. In current practice, when fat in the body is quantified via images, it is typically assessed at one slice (e.g., L4-L5) through the abdomen. This ubiquitous practice opens several questions: At what single slice anatomic location do the areas of Subcutaneous Adipose Tissue (SAT) and Visceral Adipose Tissue (VAT) estimated from a single slice best represent the corresponding volume measures? How does one ensure that the slice used for single slice area estimation from different subjects is at the homologous anatomic location? How can fat (SAT and VAT as separate components) be quantified throughout the body from low-dose CT and MR images for each body region separately?
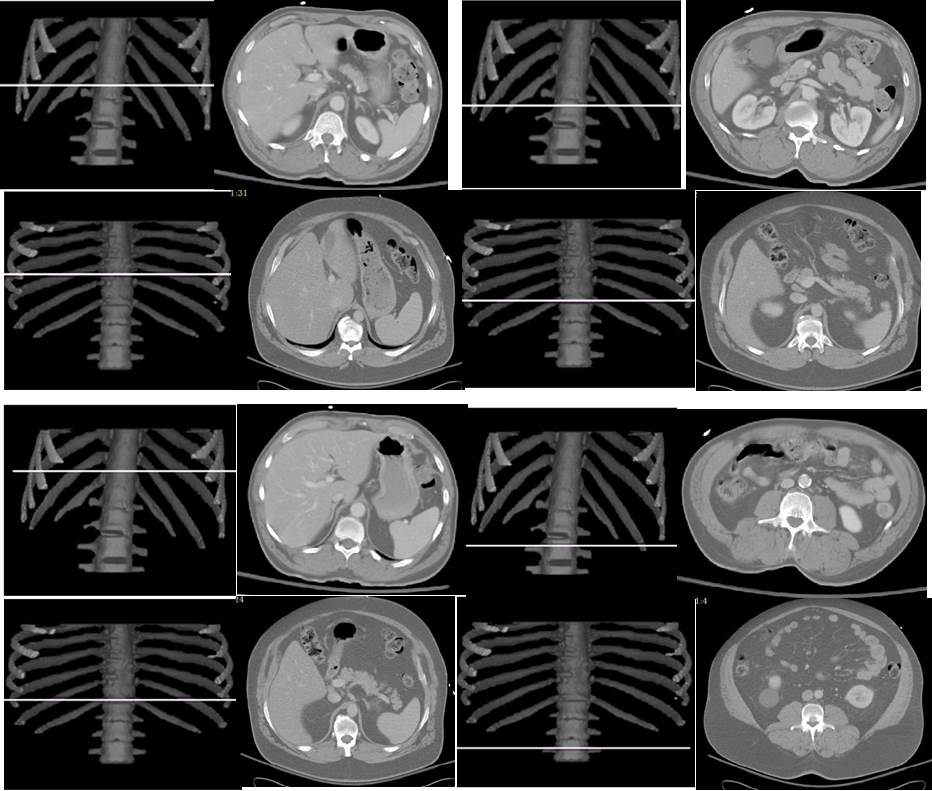
Based on AAR algorithms and the concept of standardized anatomic space, we are developing methods to answer the above questions. Our current results indicate that the best slice location (in terms of area-to-volume correlation) for SAT and VAT assessment in the abdomen is not at L4-L5, but rather at T12 for SAT and L3-L4 for VAT. Similarly, in the thorax, the best locations are at T5-T6 for SAT and T9 for VAT. The following example illustrates the best slice locations found without standardizing (top two rows – left column for SAT and right column for VAT) and with standardized anatomic space (bottom two rows – left column for SAT and right column for VAT). The two rows represent two different subjects.

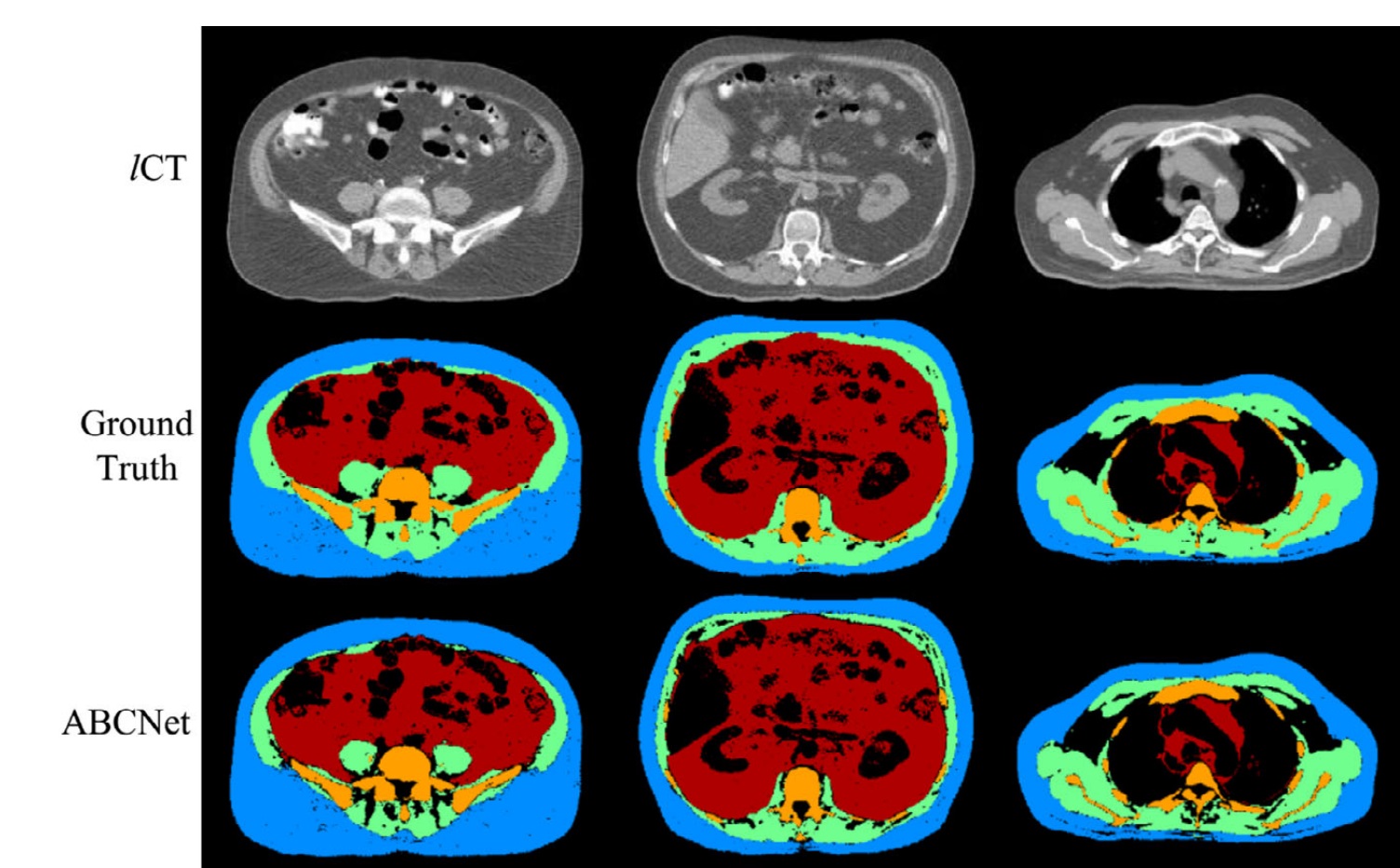
Quantification of body tissue composition is important for research and clinical purposes. In this work, we aim to automatically segment four key body tissues of interest, namely subcutaneous adipose tissue, visceral adipose tissue, skeletal muscle, and skeletal structures from body-torso-wide low-dose computed tomography (CT) images. Based on the idea of residual Encoder–Decoder architecture, a novel neural network design named ABCNet is proposed. The proposed system makes full use of multiscale features from four resolution levels to improve the segmentation accuracy. This network is built on a uniform convolutional unit and its derived units, which makes the ABCNet easy to implement. Several parameter compression methods, including Bottleneck, linear increasing feature maps in Dense Blocks, and memory-efficient techniques, are employed to lighten the network while making it deeper. The strategy of dynamic soft Dice loss is introduced to optimize the network in coarse-to-fine tuning. The proposed segmentation algorithm is accurate, robust, and very efficient in terms of both time and memory. A dataset composed of 38 low-dose unenhanced CT images, with 25 male and 13 female subjects in the age range 31–83 yrs and ranging from normal to overweight to obese, is utilized to evaluate ABCNet. We compare four state-of-the-art methods including DeepMedic, 3D U-Net, VNet, Dense V-Net, against ABCNet on this dataset. We employ a shuffle-split fivefold cross-validation strategy: In each experimental group, 18, 5, and 15 CT images are randomly selected out of 38 CT image sets for training, validation, and testing, respectively. The commonly used evaluation metrics — precision, recall, and F1-score (or Dice) — are employed to measure the segmentation quality. The results show that ABCNet achieves superior performance in accuracy of segmenting body tissues from body-torso-wide low-dose CT images compared to other state-of-the-art methods, reaching 92– 98% in common accuracy metrics such as F1-score. ABCNet is also time-efficient and memory-efficient. It costs about 18 hr to train and an average of 12 sec to segment four tissue components from a body-torso-wide CT image, on an ordinary desktop with a single ordinary GPU. Motivated by applications in body tissue composition quantification on large population groups, our goal in this study was to create an efficient and accurate body tissue segmentation method for use on body-torso-wide CT images. The proposed ABCNet achieves peak performance in both accuracy and efficiency that seems hard to improve upon. The experiments performed demonstrate that ABCNet can be run on an ordinary desktop with a single ordinary GPU, with practical times for both training and testing, and achieves superior accuracy compared to other state-of-the art segmentation methods for the task of body tissue composition analysis from low-dose CT images.
References:
1. Liu T, Pan J, Torigian DA, Xu P, Miao Q, Tong Y, Udupa JK. ABCNet: A new efficient 3D dense-structure network for segmentation and analysis of body tissue composition on body-torso-wide CT images. Med Phys. 2020;47(7):2986-99. doi: 10.1002/mp.14141. PubMed PMID: 32170754.
2. Tong Y, Udupa JK, Torigian DA. Optimization of abdominal fat quantification on CT imaging through use of standardized anatomic space: a novel approach. Med Phys. 2014;41(6):063501. doi: 10.1118/1.4876275. PubMed PMID: 24877839; PMCID: PMC4032419.